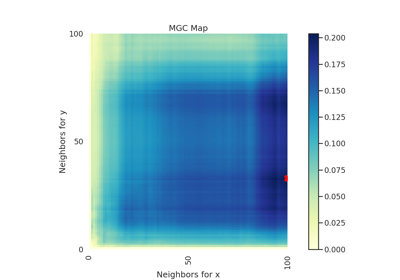
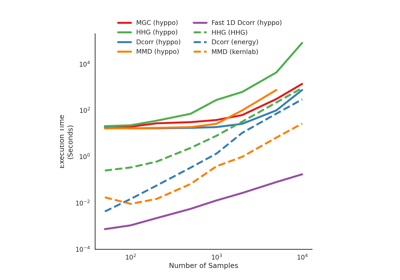
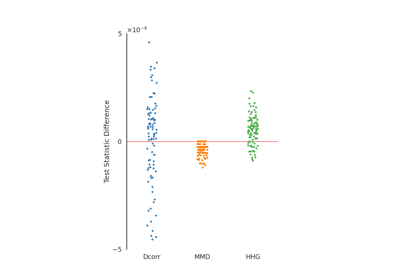
Dcorr¶
-
class
hyppo.independence.Dcorr(compute_distance='euclidean', bias=False, **kwargs)¶ Distance Correlation (Dcorr) test statistic and p-value.
Dcorr is a measure of dependence between two paired random matrices of not necessarily equal dimensions. The coefficient is 0 if and only if the matrices are independent. It is an example of an energy distance.
The statistic can be derived as follows:
Let \(x\) and \(y\) be \((n, p)\) samples of random variables \(X\) and \(Y\). Let \(D^x\) be the \(n \times n\) distance matrix of \(x\) and \(D^y\) be the \(n \times n\) be the distance matrix of \(y\). The distance covariance is,
\[\mathrm{Dcov}^b_n (x, y) = \frac{1}{n^2} \mathrm{tr} (D^x H D^y H)\]where \(\mathrm{tr} (\cdot)\) is the trace operator and \(H\) is defined as \(H = I - (1/n) J\) where \(I\) is the identity matrix and \(J\) is a matrix of ones. The normalized version of this covariance is distance correlation [1] and is
\[\mathrm{Dcorr}^b_n (x, y) = \frac{\mathrm{Dcov}^b_n (x, y)} {\sqrt{\mathrm{Dcov}^b_n (x, x) \mathrm{Dcov}^b_n (y, y)}}\]This is a biased test statistic. An unbiased alternative also exists, and is defined using the following: Consider the centering process where \(\mathbb{1}(\cdot)\) is the indicator function:
\[C^x_{ij} = \left[ D^x_{ij} - \frac{1}{n-2} \sum_{t=1}^n D^x_{it} - \frac{1}{n-2} \sum_{s=1}^n D^x_{sj} + \frac{1}{(n-1) (n-2)} \sum_{s,t=1}^n D^x_{st} \right] \mathbb{1}_{i \neq j}\]and similarly for \(C^y\). Then, this unbiased Dcorr is,
\[\mathrm{Dcov}_n (x, y) = \frac{1}{n (n-3)} \mathrm{tr} (C^x C^y)\]The normalized version of this covariance [2] is
\[\mathrm{Dcorr}_n (x, y) = \frac{\mathrm{Dcov}_n (x, y)} {\sqrt{\mathrm{Dcov}_n (x, x) \mathrm{Dcov}_n (y, y)}}\]The p-value returned is calculated using a permutation test using
hyppo.tools.perm_test. The fast version of the test useshyppo.tools.chi2_approx.When the data is 1 dimension and the distance metric is Euclidean, and even faster version of the algorithm is run (computational complexity is \(\mathcal{O}(n \log n)\)) [3].
- Parameters
compute_distance (
str,callable, orNone, default:"euclidean") -- A function that computes the distance among the samples within each data matrix. Valid strings forcompute_distanceare, as defined insklearn.metrics.pairwise_distances,From scikit-learn: [
"euclidean","cityblock","cosine","l1","l2","manhattan"] See the documentation forscipy.spatial.distancefor details on these metrics.From scipy.spatial.distance: [
"braycurtis","canberra","chebyshev","correlation","dice","hamming","jaccard","kulsinski","mahalanobis","minkowski","rogerstanimoto","russellrao","seuclidean","sokalmichener","sokalsneath","sqeuclidean","yule"] See the documentation forscipy.spatial.distancefor details on these metrics.
Set to
Noneor"precomputed"ifxandyare already distance matrices. To call a custom function, either create the distance matrix before-hand or create a function of the formmetric(x, **kwargs)wherexis the data matrix for which pairwise distances are calculated and**kwargsare extra arguements to send to your custom function.bias (
bool, default:False) -- Whether or not to use the biased or unbiased test statistics.**kwargs -- Arbitrary keyword arguments for
compute_distance.
Methods Summary
|
Helper function that calculates the Dcorr test statistic. |
|
Calculates the Dcorr test statistic and p-value. |
-
Dcorr.statistic(x, y)¶ Helper function that calculates the Dcorr test statistic.
- Parameters
x,y (
ndarray) -- Input data matrices.xandymust have the same number of samples. That is, the shapes must be(n, p)and(n, q)where n is the number of samples and p and q are the number of dimensions. Alternatively,xandycan be distance matrices, where the shapes must both be(n, n).- Returns
stat (
float) -- The computed Dcorr statistic.
-
Dcorr.test(x, y, reps=1000, workers=1, auto=True, perm_blocks=None)¶ Calculates the Dcorr test statistic and p-value.
- Parameters
x,y (
ndarray) -- Input data matrices.xandymust have the same number of samples. That is, the shapes must be(n, p)and(n, q)where n is the number of samples and p and q are the number of dimensions. Alternatively,xandycan be distance matrices, where the shapes must both be(n, n).reps (
int, default:1000) -- The number of replications used to estimate the null distribution when using the permutation test used to calculate the p-value.workers (
int, default:1) -- The number of cores to parallelize the p-value computation over. Supply-1to use all cores available to the Process.auto (
bool, default:True) -- Automatically uses fast approximation when n and size of array is greater than 20. IfTrue, and sample size is greater than 20, thenhyppo.tools.chi2_approxwill be run. Parametersrepsandworkersare irrelevant in this case. Otherwise,hyppo.tools.perm_testwill be run. Ifxandyhave p equal to 1 andcompute_distanceset to'euclidean', then and \(\mathcal{O}(n \log n)\) version is run.perm_blocks (
Noneorndarray, default:None) -- Defines blocks of exchangeable samples during the permutation test. If None, all samples can be permuted with one another. Requires n rows. At each column, samples with matching column value are recursively partitioned into blocks of samples. Within each final block, samples are exchangeable. Blocks of samples from the same partition are also exchangeable between one another. If a column value is negative, that block is fixed and cannot be exchanged.
- Returns
Examples
>>> import numpy as np >>> from hyppo.independence import Dcorr >>> x = np.arange(25) >>> y = x >>> stat, pvalue = Dcorr().test(x, y) >>> '%.1f, %.2f' % (stat, pvalue) '1.0, 0.00'
In addition, the inputs can be distance matrices. Using this is the, same as before, except the
compute_distanceparameter must be set toNone.>>> import numpy as np >>> from hyppo.independence import Dcorr >>> x = np.ones((10, 10)) - np.identity(10) >>> y = 2 * x >>> dcorr = Dcorr(compute_distance=None) >>> stat, pvalue = dcorr.test(x, y) >>> '%.1f, %.2f' % (stat, pvalue) '0.0, 1.00'